Get to know the clustering tiling: knowing more about the minority (the rare cells) with a little help from the majority
Cells may also disappear en masse. For example, in the tissues that line the inner walls of the small intestine, the liver or the kidneys, there are unique types of cells whose number is dwarfed compared to their more common neighbors. However, despite their small numbers, these cells play a significant role in maintaining our health. Recently, Weizmann Institute of Science scientists have developed A new method for genetic sequencing which allows to trace rare cells in a wide variety of tissues. The method that has been named "Clump Sequencing" (Clump Sequencing or ClumpSeq for short) makes it possible to quickly and relatively easily locate cells that have so far managed to escape the existing sequencing technologies, and to characterize their various functions depending on their location in the tissue.
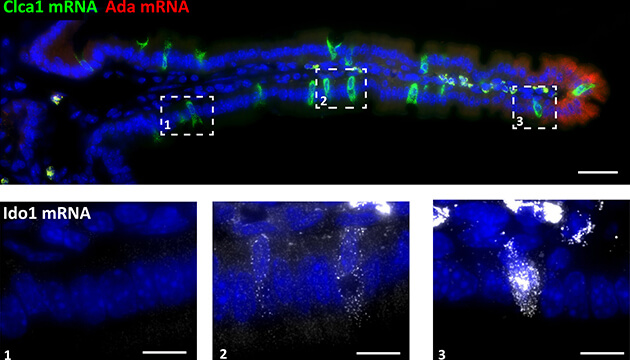
90% of the inner wall of the small intestine consists of food-absorbing cells called enterocytes. As important as they are, understanding the functioning of the intestine will not be complete without also knowing the goblet cells, which although make up less than 10% of the inner wall, but are exclusively responsible for the secretion of the mucous material required for the protection of the intestine, or the enteroendocrine cells which make up less than 1%, but secrete hormones that are essential for metabolic processes, such as control of sugar levels in the body. The methods that exist today make it possible to sequence the genetic material at the level of the single cell - and not only at the level of the tissue - but rare cells pose a challenge: for example, when sequencing the genetic material of cells in the intestine, the RNA content of the rare cells is XNUMX% null in the data flow of Their neighbors are the enterocytes.
However, despite the fact that the common cells screen and eliminate the rare cells - in the laboratory of Prof. His Itzkowitz From the department of molecular biology of the cell, they realized that they can also help locate their neighbors hiding in the dishes. in a previous study They showed in Prof. Itzkovitz's laboratory how, during their short life, enterocytes frequently change their nature and function according to their location in the intestine - similar to factory workers changing tasks along the production line. They are formed at the base of the villi - tiny finger-like protrusions that increase the surface area of the intestine 30 times - migrate along them, and finally fall to their death at the ends of the "fingers". The entire process takes about four days, during which the mice adjusted their genetic expression patterns depending on their position on the roots. These changes in the patterns of genetic expression, Prof. Itzkowitz thought, could help detect the rare cells.
In the new method they developed, as its name implies, the scientists first divide the tissue into clusters, with each cluster containing a limited number of cells - between two and ten; This division in itself increases the chance of finding rare cells compared to the chance of finding them in tissue samples containing hundreds of cells. Next, the scientists sequence the messenger RNA molecules of each cluster and scan the resulting RNA pool to locate genetic "landmarks" of the common cells - in this case the enterocytes. These landmarks are similar to geographic landmarks used for navigation - except that instead of a street lamp or a monument, they are genetic expression patterns associated with one or another area of the tissue. The landmarks of the common cells make it possible to infer the original location of each of the clusters, on the rare cells in them, and then search accordingly for corresponding genetic landmarks of the rare cells depending on their location in the tissue.
When the researchers applied the method to mouse intestine samples, they discovered a wealth of new details about the cells in this tissue: the goblet cells, they learned, change their function almost like the enterocytes. The expression patterns of about a third of all their genes changed as they moved along the roots. It was also discovered that genes involved in the control of the immune system were mostly expressed only at the tip of each cyst. "Because the end of the hilum is an area rich in intestinal bacteria, and it is also the point from which the enterocytes fall to their death and thus burst a microscopic opening in the tissue," explains Prof. Itzkowitz, "but it makes sense that cells located in this area would be involved to one degree or another in immune control processes to prevent the development of inflammation Unnecessary". In addition, they revealed that certain types of hormone-secreting cells, which were previously thought to also move along the cilia, are actually stationary: they appear at the base of every cilia - and stay there.
But we are not only dealing with intestines. The cluster sequencing method is expected to make it possible to characterize the activity of rare cells in different tissues, and how this activity goes awry in rarer and less rare diseases. For example, a rare type of cells called tuft cells are known for their involvement in immune control processes in the gut and lungs; Characterizing their activity depending on their exact location in the tissue could help to better understand how allergies and inflammatory bowel diseases develop. The new method can also be mobilized to fight cancer - as it allows to track changes in the activity of cancer cells depending on their location in the tumor.
Post-doctoral researchers Dr. Rita Manko and Dr. Ina Averbuch and faculty scientist Dr. Karen Behar Halpern also participated in the study - all from Prof. Itzkovitz's group. Dr. Ziv Porat from the Department of Life Science Research Infrastructures and Prof. Ido Amit from the Department of Immunology also participated.
