The new technology - DBlink - is based on the SMLM method and an artificial intelligence algorithm (neural networks) and makes it possible to reproduce fast dynamics of biological objects in super-resolution in living cells
Researchers at the Faculty of Biomedical Engineering at the Technion have developed a system forTracking dynamic processes in living cells. the research published in the journal Nature Methods Led by Prof. Yoav Shechtman and PhD student Alon Sagi from the Faculty of Biomedical Engineering. The new technology - DBlink - is based on the SMLM method and an artificial intelligence algorithm (neural networks) and makes it possible to reproduce fast dynamics of biological objects in super-resolution in living cells.
One of the most important challenges in microscopy is monitoring dynamic biological processes with high resolution (separation) in time and space. Conventional optical microscopes are limited by the diffraction limit, which limits the resolution of the microscope to about half the wavelength of light. This means that in visible light, the diffraction limit is around 300-200 nm. Therefore we will not be able to see many of the objects of interest to researchers in the field of biology, for example certain viruses (100 nm), proteins (10 nm) and DNA molecules (2.5 nm).
The diffraction limit was first formulated by the German physicist Ernst Carl Abbe in 1873. Abbe's determination, according to which the resolution of an optical microscope is limited to about half the wavelength, occupied many researchers and engineers, and in recent years new technologies have succeeded in "circumventing" the same limitation and providing super-resolution - a separation of Tens of nanometers, and in some cases even single nanometers.
One of the breakthroughs in this regard is the development of a method for the detection of single molecules (SMLM) by Eric Betzig, who was awarded the Nobel Prize in Chemistry for this in 2014 together with William Morner and Stefan Hell. This method is based on the connection of fluorescent (luminous) molecules to the biological structure. It makes it possible to accurately locate the location of the molecules, which indicates the biological structure being photographed. The method revolutionized biological imaging and improved the spatial resolution by an order of magnitude relative to conventional methods.
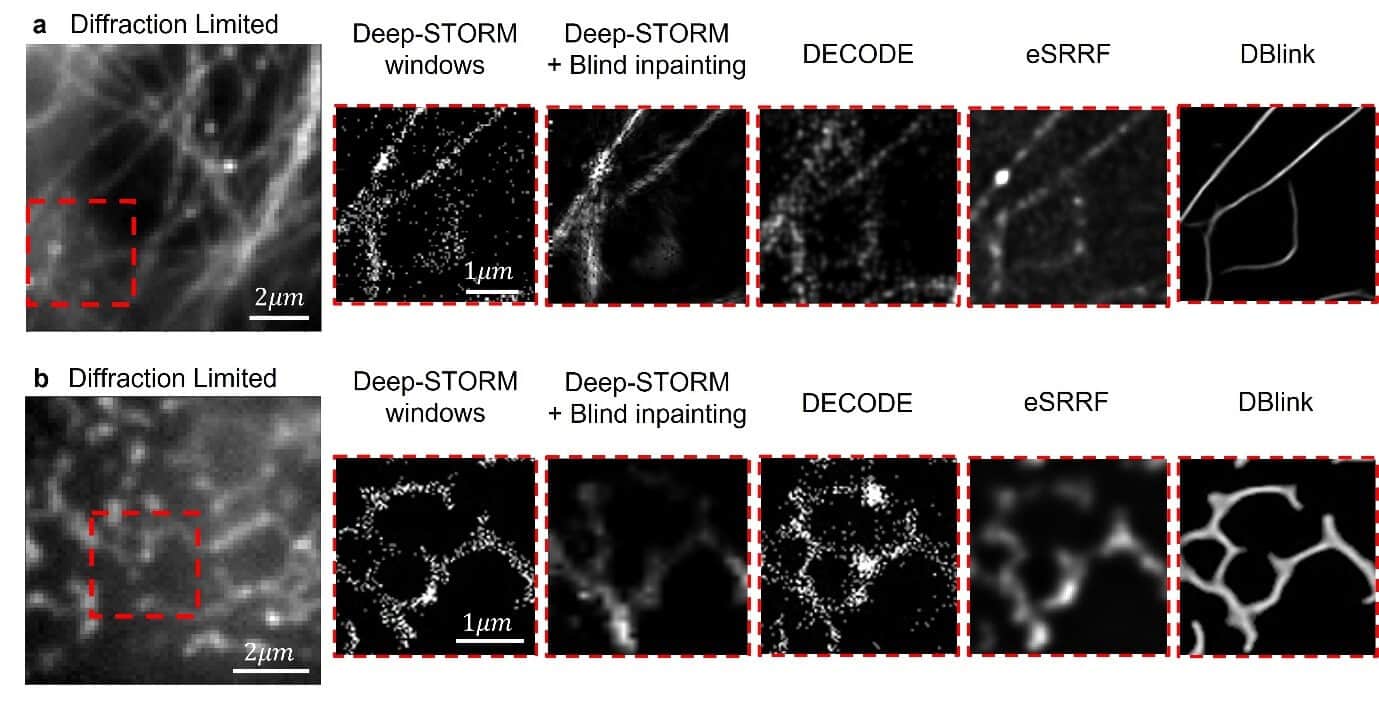
However, the method requires a long shooting time to improve the spatial resolution and does not allow dynamic object tracking. Since most biological processes occur in a dynamic environment in living cells, researchers were looking for a solution that would allow this.
One of the solutions to the resolution problem is the use of short waves, because the resolution is limited by the wavelength and therefore short waves will allow a higher resolution. Another option is to perform a series of scans and analyze them afterwards. However, both of these options have significant disadvantages in the biological context: short waves are high-energy waves that damage the living cell during photography, and performing a long series of scans prolongs the duration of photography and is not suitable for the dynamics of living cells that are in constant motion.
This is where DBlink comes into play - the new technology presented by Sagi and Prof. Shechtman in their article. The demonstration of the new technology was conducted on several organelles in living cells, for example mitochondria - the energy producer of the living cell. Using the new technology, the mitochondrial structure and its natural dynamics were reconstructed in super resolution. The researchers demonstrated a spatial resolution of 30 nm and a time resolution of 15 hundredths of a second. One of the distinct advantages of the method is the ability to weigh the movement of biological objects and produce an accurate and clear image, even though the object under examination moves during the photograph.
The study was supported by the German Research Foundation, the European Union (through the Horizon 2020 program) and the Zuckerman Foundation.
for the article in the journalNature Methods
For a video showing the new technology click here
On the left - the video taken with the microscope. On the right - the video edited by DBlink and showing the movement of the mitochondria
For pictures click here
- Prof. Yoav Shechtman
- Alon Sagi
Photo credit: Technion spokespeople

2 תגובות
All microscopes have a resolution limit that determines the size of the smallest objects that can be seen in their medium. In recent years, computational methods have been developed that allow overcoming this limitation and achieving super-resolution (beyond the original limitation). However, these methods require a long imaging time to obtain a single super-resolution image. This compromise is a problem in biological research, because it allows observing only static (dead) cells and does not allow monitoring of cellular activity. As you might expect, a significant part of cell research requires the ability to image activity in live, dynamic cells. DBlink, the method presented in this work, makes it possible to observe cellular activity in super resolution and thus allows researchers to see and understand processes that previously could not be studied.
There is an explanation of the problem, not of the solution. So what is this method, DBLink, actually?