A dramatic achievement in the study of the ubiquitin system: resolved spatial structure of a triubiquitin chain that binds to a protein. The research was conducted at the Technion and Stanford University
Research conducted at the Technion and Stanford University, and now published in the journal Structure, reveals for the first time the structure of triple-repeat ubiquitin chains. The research group at the Technion was led by Prof. Ariel Stanhill, a faculty member at the Rappaport Faculty of Medicine at the Technion.
The function of proteins in the living cell depends on their proper folding. Disrupted folding (proteotoxicity) may damage the cell, and subsequently cause damage at the macro level, that is, in the body tissues and the entire body. Such damages are involved in a wide spectrum of diseases, including neurodegenerative diseases such as Alzheimer's.
The main defense system that prevents those damages is the ubiquitin mechanism, the discovery of which led to the winning of Professors Avraham Hershko and Aharon Chachanover from the Technion and their colleague Prof. Irwin Rose from the University of California with the Nobel Prize in Chemistry (2004). The ubiquitin system marks the damaged proteins with a "death tag" made of ubiquitin chains, and this tagging leads to its removal to the proteasome, the "cellular trash can". Later it became clear that this system is involved in other biological processes.
The discovery by Hershko, Chechenover and Rose opened a new field of research in science and medicine: controlled processes of protein breakdown in the cell. Now, with the publication of Prof. Stanhill's research, a dramatic step has been taken in this field: deciphering the three-dimensional structure of complex ubiquitin chains (tri-ubiquitin chains).
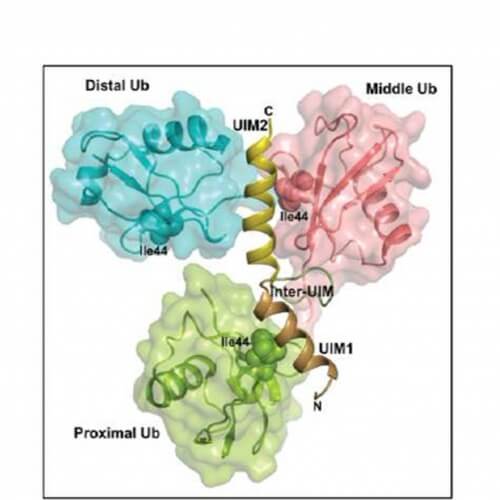
In an article published in Structure, Prof. Stanhill and his colleagues report on the solution of the spatial structure of the AIRAPL protein and of a three-protein chain of ubiquitin proteins. The group discovered for the first time how this chain is distinguished from other ubiquitin chains, and revealed the links in this system. According to Prof. Stanhill, "This is the first description of complex ubiquitin chain binding. We were able to show that a protein called AIRAPL binds two ubiquitin molecules through a unique interaction surface (UIM2), and at the same time enables another link on the ubiquitin receptor. This enables a 'triple bond' leading to the launch of the damaged protein for degradation in the proteasome. In our estimation, the solved structure will allow prediction of the binding potential of similar ubiquitin chains.'
