Natural selection: Technion researchers have uncovered an evolutionary mechanism that protects the organism from intracellular interactions that endanger it. Possible application: targeted damage to hostile bacteria that attack humans
The scientific journal PNAS reports the discovery of an evolutionary mechanism that protects bacteria and other organisms from molecular interactions that endanger them. This unique mechanism, discovered by the research team of Prof. Noam Adir from the Technion, protects all living things by filtering out "lethal sequences" of amino acids in proteins that are going to harm the organism. The use of information about the mechanism will allow precise damage to harmful bacteria without harming the person carrying them.
Evolution, as we know, is a process in which mutations - changes in DNA, the genetic material of the organism, are preserved or disappear according to the survival advantage they give to the organism. An organism that will be more successful - in the sense of adapting to the environment and producing fertile offspring - will enjoy better chances of surviving and passing on its traits to future generations. It is worth noting in this context that the phrase "survival of the fittest" is not a correct scientific wording, since the stronger individual is not necessarily the individual better suited to the environment. The correct formulation, as coined by Charles Darwin, is "survival of the fittest".
In the process of natural selection, which is one of the basic principles of evolution, the competing individuals are exposed to evolutionary pressure, which is the set of factors that affect the individual's adaptation to the environment: resistance to diseases, predators, climatic challenges, etc. and the ability to produce fertile offspring that will survive and pass on those environment-compatible traits .
An innovative method
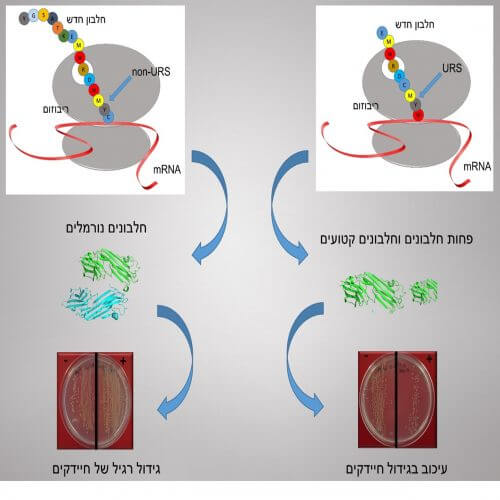
The research published in PNAS was conducted by Prof. Adir, a faculty member at the Shulich Faculty of Chemistry at the Technion, together with Dr. Sharon Panias-Navon and Master's student Tali Schwartzman from his research group. The researchers used an innovative method to identify URSs - underrepresented sequences, i.e. missing or rare sequences, from the vast database of biological information. After identifying these sequences with the method, they tested, experimentally, the effect of URSs on bacteria. The conclusion: the rarity of specific sequences in a specific bacterium is derived from the potential damage of these sequences, which may inhibit the creation of proteins (synthesis) and inhibit the development of the bacterium. In other words, the evolutionary pressure reduces, over time, the frequency of "lethal sequences" that threaten the resilience of the bacterium.
"The molecular machines that make life possible," explains Prof. Adir, "are long polymers built from linear sequences of different chemical groups: proteins, DNA and RNA. The incredible diversity that characterizes the biological world is a consequence of the evolutionary changes applied to these polymers."
At the molecular level, this variation mainly stems from the astronomical number of different possibilities for the arrangement of the natural amino acids from which the proteins are built. This number is derived from the existence of 20 natural amino acids; Using just three of them leads to 8,000 different possible sequences, using five allows more than 3 million different sequences, and so on. A typical protein does not have three or five amino acids but hundreds. "The amino acid sequence determines the three-dimensional structure of the protein and hence its characteristics and abilities. The binding of the protein to other proteins may create new and improved abilities, but a wrong connection may damage the protein and even lead to the death of the cell, i.e. the bacterium."
Such incorrect connections can result from the random formation of "negative" mutations, i.e. those that are not expected to survive because they reduce the organism's ability to survive in the environment. This is where one of the most important scientific revolutions of recent years comes into the picture - bioinformatics, in the framework of which enormous information is accumulated on the DNA sequences of entire organisms - from simple bacteria to humans. "The current research resulted from the understanding that this database provides us with an opportunity to check not only the common sequences in various organisms, as many research groups do in the world, but also the URSs - the missing and rare sequences."
To this end, the researchers wrote a computer program that scans all the coded sequences for all possible proteins (the proteomes) in the genomes of various microorganisms, mainly pathogenic (causing diseases). The goal: detection of URSs in specific organisms. profile
The URSs vary from bacterium to bacterium, and at this stage of the study the researchers focused on bacteria E. coli.
The experiment: transplanting rare sequences
Now the researchers sought to prove that the URSs are indeed harmful (hence their rarity) and to determine the exact place of their injury. To do this, they grafted these URS sequences into normal proteins. The result, which can be seen on the right side of the attached diagram: the implantation of the URS damaged the synthesis process of new proteins. Furthermore, when longer URS sequences were used - a sequence of 4 amino acids - the presence of the URS sequence led to the inhibition of the development of the bacteria and even to its death.
Combining the amino acids to form a new protein is carried out by the ribosome, which receives the instructions from the genetic code. Therefore, the researchers turned to experts who specialize in measuring the activity of individual ribosomes: Prof. Joseph Puglisi from Stanford University and his doctoral student, Guy Kornberg. Puglisi and Kornberg also implanted the URS sequences in the protein and confirmed the findings: these sequences indeed inhibit the translation process and the development of the protein. Furthermore, they determined exactly where in the ribosome the delay in the translation process occurs: at the entrance to the exit channel of the ribosome, from which the new proteins "emerge".
The research group at the Technion even implanted the same sequences in the protein in a culture of human cells, and the result: no effect on the protein was evident, meaning no harm was caused to the person. "These findings," says Prof. Adir, "make it clear that the use of URSs may target the bacteria we want to kill, without harming the person in whose body they are. In light of this, and with the hope that in the near future the practical possibilities of our discovery will be tested, we registered a patent for it together with the Technion."
